Inhaled allergen
Mite allergens. Our group is working on several inhaled allergens. For example, we are studying Group 1 and Group 2 dust mite allergens (Der f 1, Der p 1, Der f 2 and Der p 2), which are believed to be among the most potent triggers of asthma. We hope that our studies, which are performed in collaboration with INDOOR Biotechnologies, Inc will ultimately lead toward an effective immunotherapy for people allergic to dust mites.
Most mite allergic patients (>80%) have IgE antibodies against the Group 1 mite allergens, Der f 1 and Der p 1. These allergens are cysteine proteases and their proteolytic activity contributes significantly to allergenicity. Der p 1 cleaves the CD23 and CD25 receptors, and the cleavage of these receptors favors a Th2 response and induction of release of pro-inflammatory cytokines from bronchial epithelial cells, mast cells and basophils. The resulting increase in IgE antibody synthesis and inflammation of lung epithelium may explain why mite allergens are strongly associated with asthma.
Food allergens
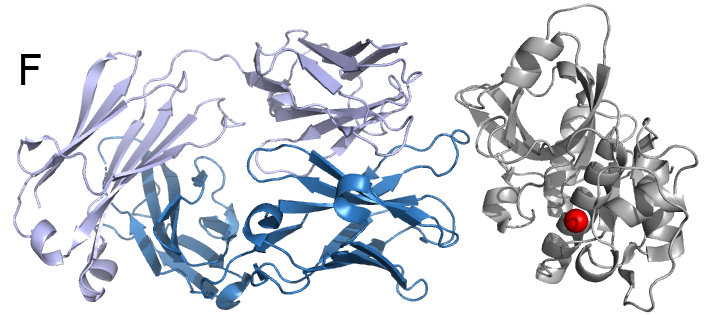
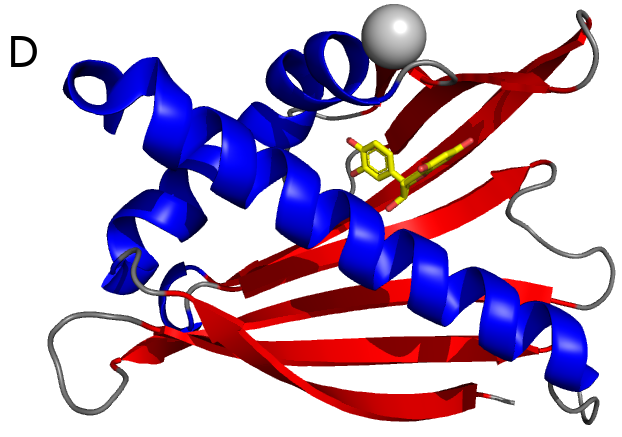
Peanut and treenut allergens. Peanut allergens alone affect 1% of the population in the US and they often cause anaphylaxis, the most dangerous—and often life-threatening—manifestation of allergic diseases. These studies are performed in collaboration with researchers from the US Department of Agriculture and institutes and universities in Europe. We are working both on recombinant allergens and those isolated from natural sources, for example Ara h 1 (Fig. C) and Ara h 8 (Fig D).
Studies of natural allergens are especially important, because in many cases it is difficult to demonstrate in recombinant proteins the same IgE binding properties that are seen in natural allergens. Differences in IgE binding properties between recombinant and natural versions of allergens may indicate that in diagnostic tests, the recombinant protein is not a suitable replacement for the allergen derived from its natural source.
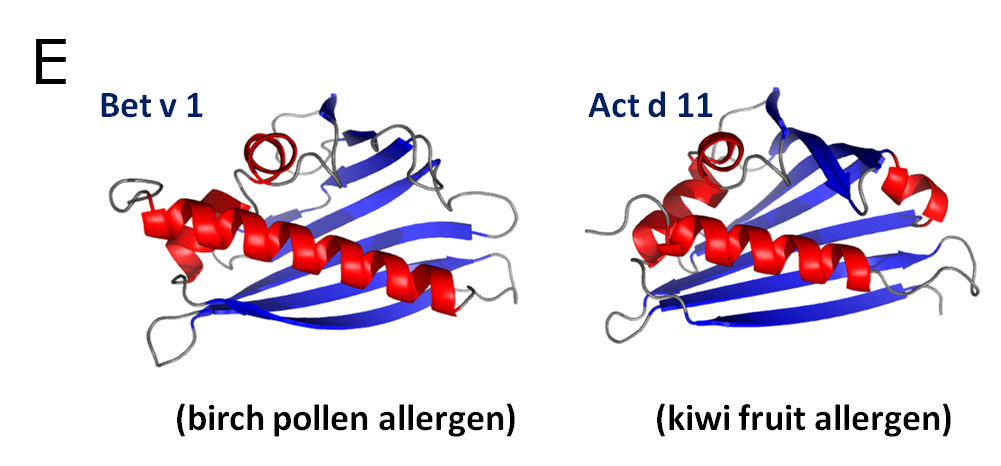
Fruit allergens. Our group is also working on several fruit allergens. One of such group of allergens, of which we are working in collaboration with Dr. Ciardiello, are kiwifruit allergens (e.g. Act d 5, Act c 10 and Act d 11). For example, the major latex protein Act d 11 (kirola) is expressed in large amounts in ripe green and gold kiwifruit. Ten percent of kiwifruit allergic individuals produce IgE that recognizes Act d 11, thus making it an allergen of high interest. Interestingly, Glu46 (Glu45 in Bet v 1), which was shown to be important for IgE binding in Bet v 1, is conserved in Act d 11, despite the fact that it is not conserved in other allergens with significantly higher sequence identity to the birch pollen allergen. It is suggested that the so called Gly-rich loop (P-loop), which is conserved in all PR-10 related allergens (Fig. E), may be responsible for the IgE cross-reactivity between Bet v 1 and Act d 11. Despite these analyses, the biological function of Act d 11 remains unknown.
Agricultural pests
Together with several laboratories from the University of South Carolina and other universities we are working on projects related to agricultural pests, such as Tetranychus urticae (two-spotted spider mite; TSSM). Our role is to provide structural and molecular data which will allow us to understand molecular basis of resistance of these species to pesticides. Moreover, we are interested in the molecular basis of pest-plant interactions. Below are some examples of the proteins that we are working on.
Many plant species are cyanogenic and are able to release toxic hydrogen cyanide (HCN) from glycoside or lipid precursors upon herbivore attack. Various studies have demonstrated that the T. urticae overcomes cyanide toxicity by expressing genes of foreign origin within its genome acquired through the process of horizontal gene transfer. In addition, it was shown that two mutually nonexclusive cyanide detoxification pathways, catalyzed by proteins coded by two horizontally transferred genes, may be present in TSSM. Cyanide might be first oxidized to cyanate (OCN−), which is then catabolized by the horizontally acquired TSSM cyanase (Tetur28g02430) into ammonia and carbon dioxide (Fig. G).
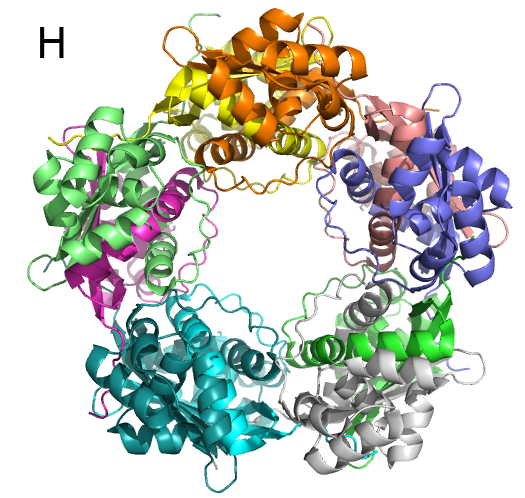
We have characterized T. urticae cyanase and determined the structure of this enzyme (Fig. H). The quaternary structure of the enzyme was determined to be decameric, and the decamer may be treated as a pentamer of dimers. Two dimers are necessary to create an active site; therefore, there are five active sites in one decamer.
In addition to proteins involved in HCN detoxification, we are interested in several protein families that help TSSM to detoxify xenobiotics and/or acaricides. For example, we are studying enzymes like P450 monooxygenases, glutathione-S-transferases, UDP-glycosyltransferases and intradiol ring-cleavage dioxygenases.
Intradiol-ring cleavage dioxygenases (ID-RCDs) cleave aromatic compounds by utilizing a non-heme iron cofactor in the active site TSSM acquired ID-RCDs by horizontal gene transfer and 17 of these proteins were identified in this agricultural pest. We have structurally and functionally characterized Tetur07g02040, and this is the first TSSM ID-RCD that was characterized on the molecular level. This is also the first structurally characterized ID-RCD that originates from a Eukaryote. The structural analysis revealed that Tetur07g02040 is monomeric (Fig. 2B), which is highly unusual for ID-RCDs.
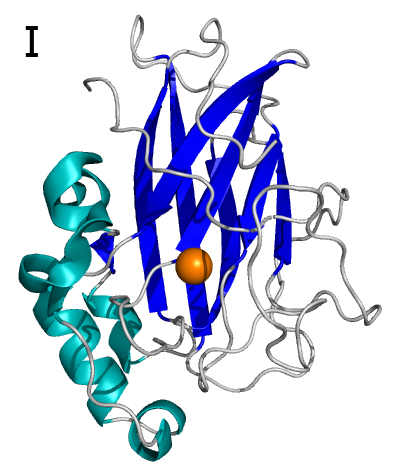
Tetur07g02040 has several unusual structural features that differentiate this enzyme from other family members that have their structures determined. Not only is the TSSM protein monomeric, but it also has a relatively open active site and an N-terminal fragment that is disordered.
Bacterial proteins
Currently our lab is working on several bacterial proteins originating from Mycobacterium tuberculosis, Neisseria gonorrhoeae, Staphylococcus aureus, Vibrio vulnificus and Wolbachia species. We are interested in proteins that are targets for potential antimicrobial compounds.
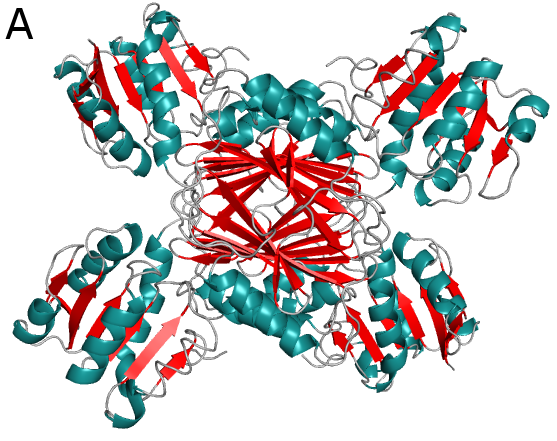
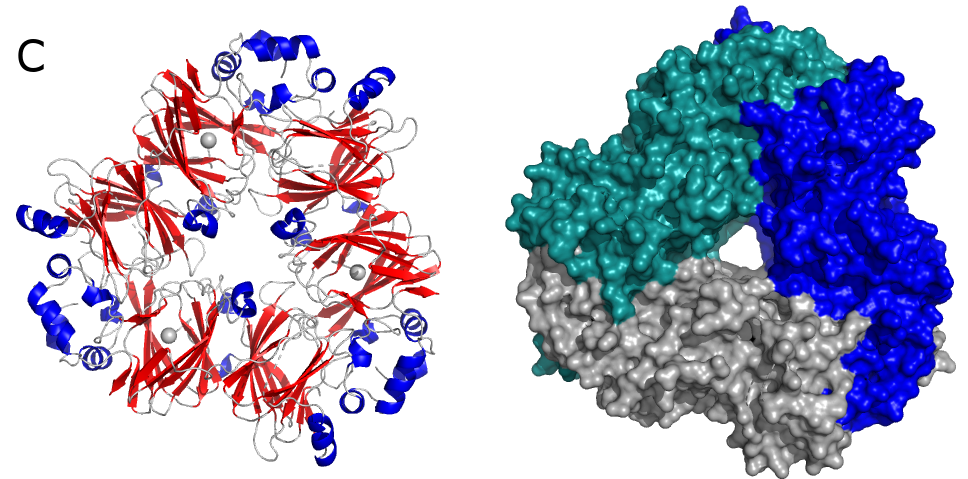
Figure above (A) shows crystal structures of 4-hydroxytetrahydrodipicolinate reductase (DapB) from M. tuberculosis. DapB is a part of the pathway leading to meso-diaminopimelate and lysine synthesis. This compounds are critical for peptidoglycan productions and survival of bacteria.
Another potential drug target is 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR), which is involved in steroid synthesis in the non-mevalonate pathway. DXRs have dimeric structures (Fig. B) and require a divalent cation (e.g. Mg2+) for their activity.
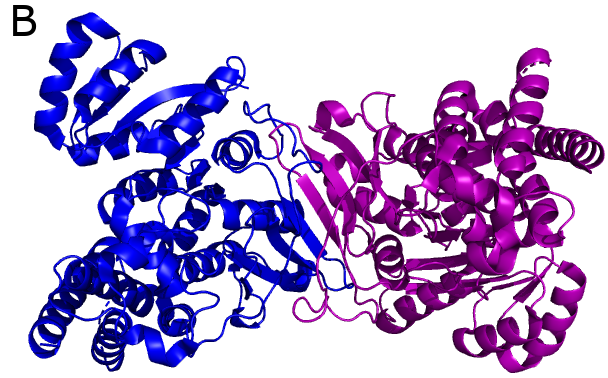
Both DapBs and DXRs are potential drug targets as they are critical parts of bacterial metabolic pathways, which are not present in humans.